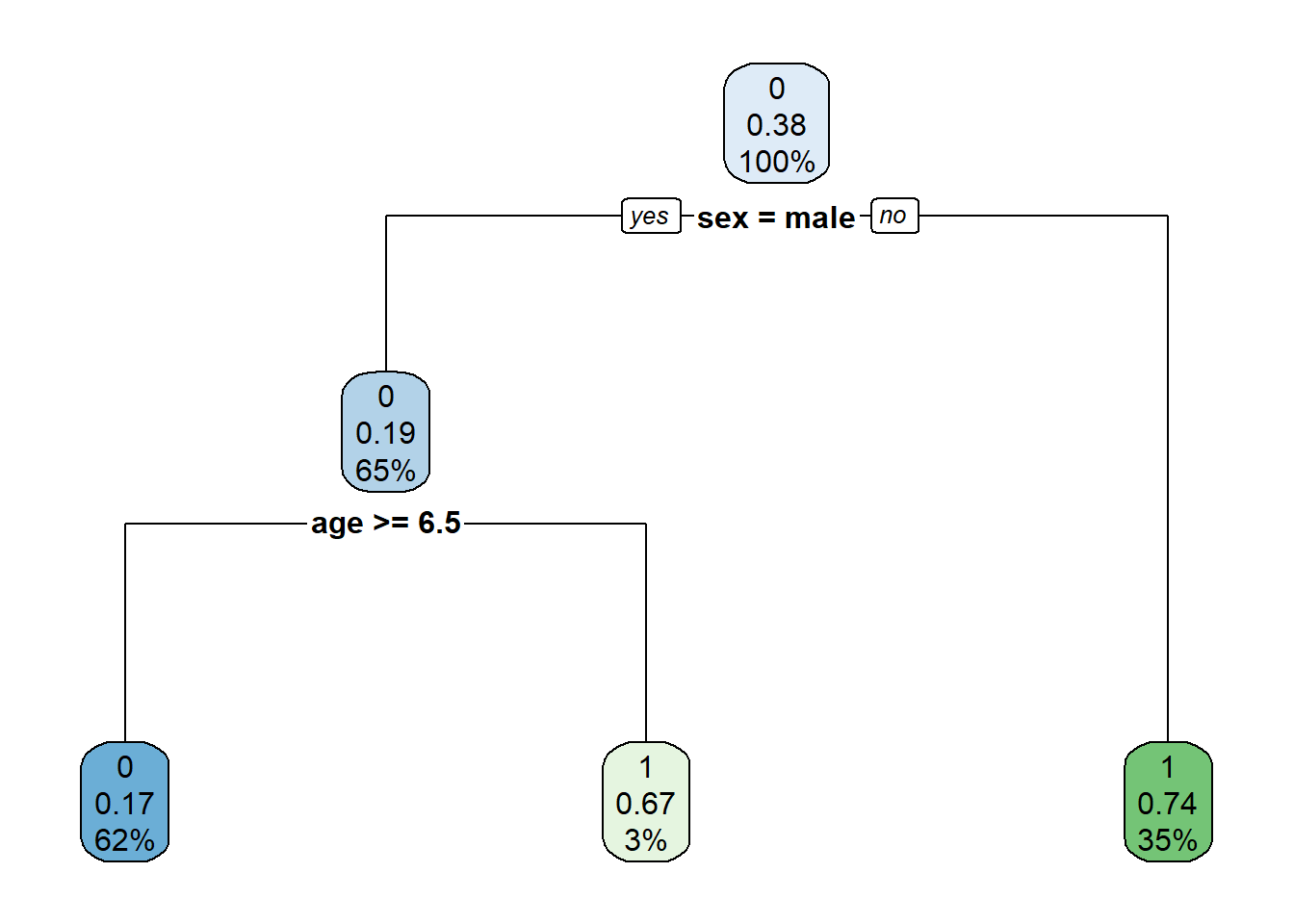
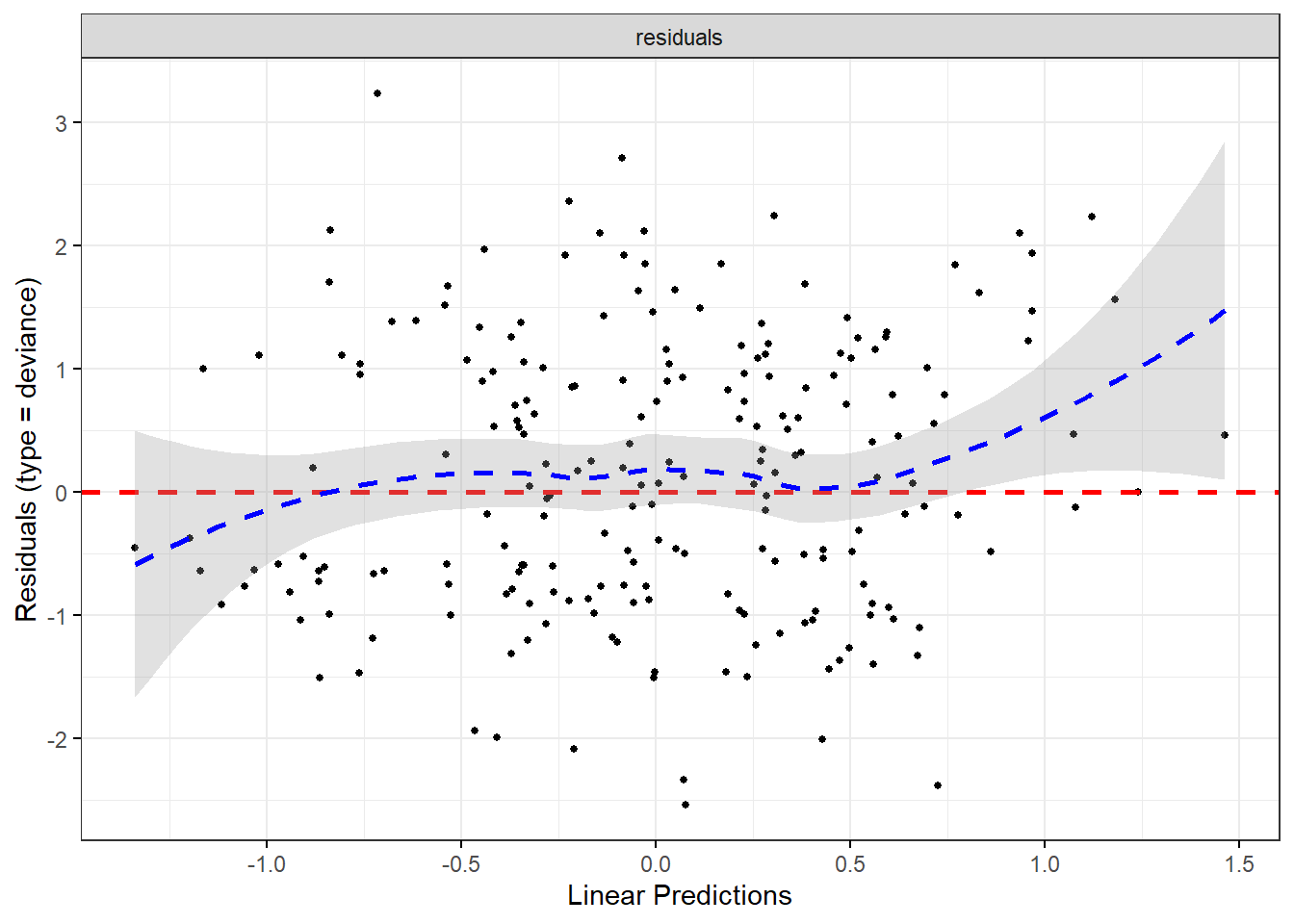
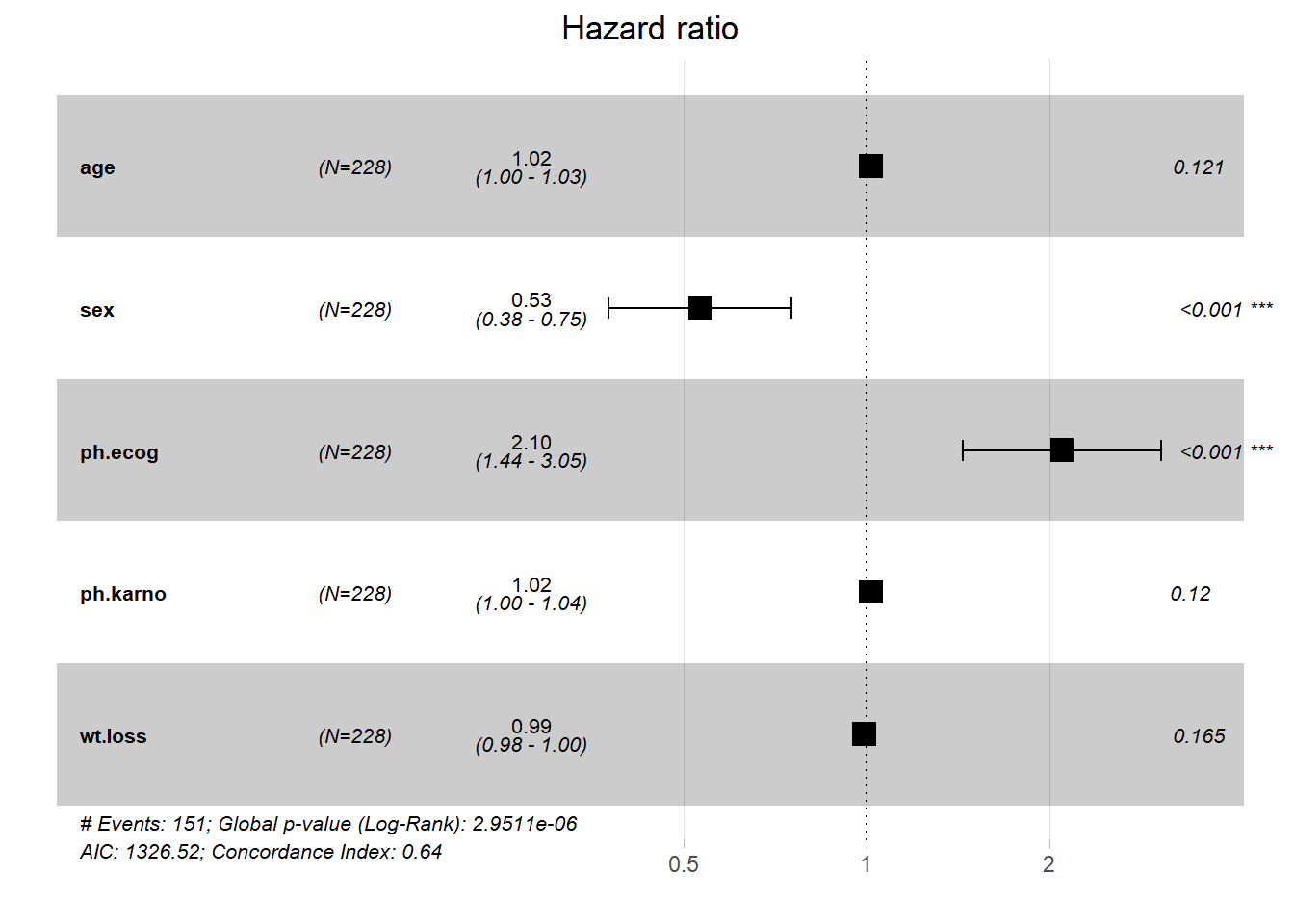
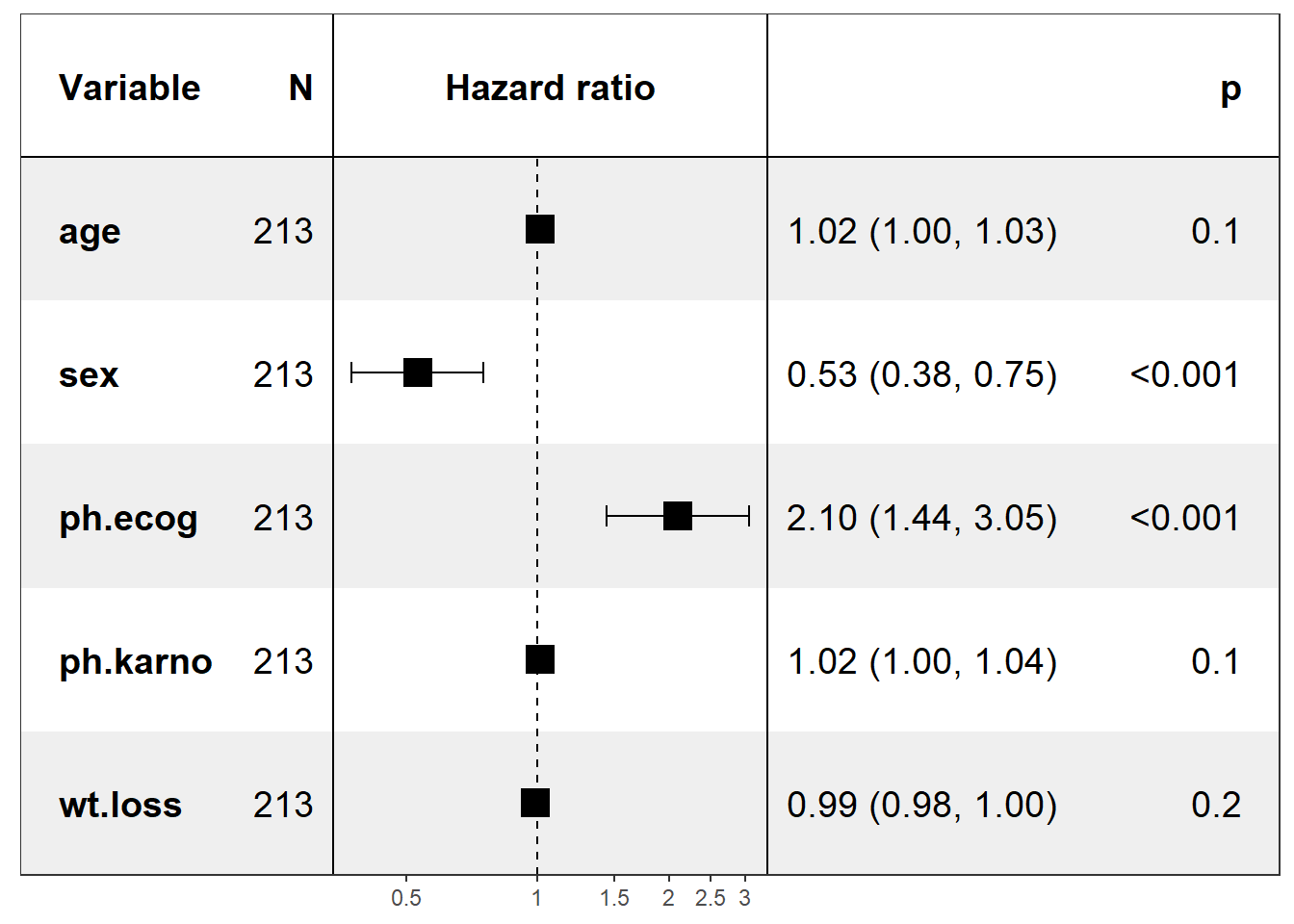
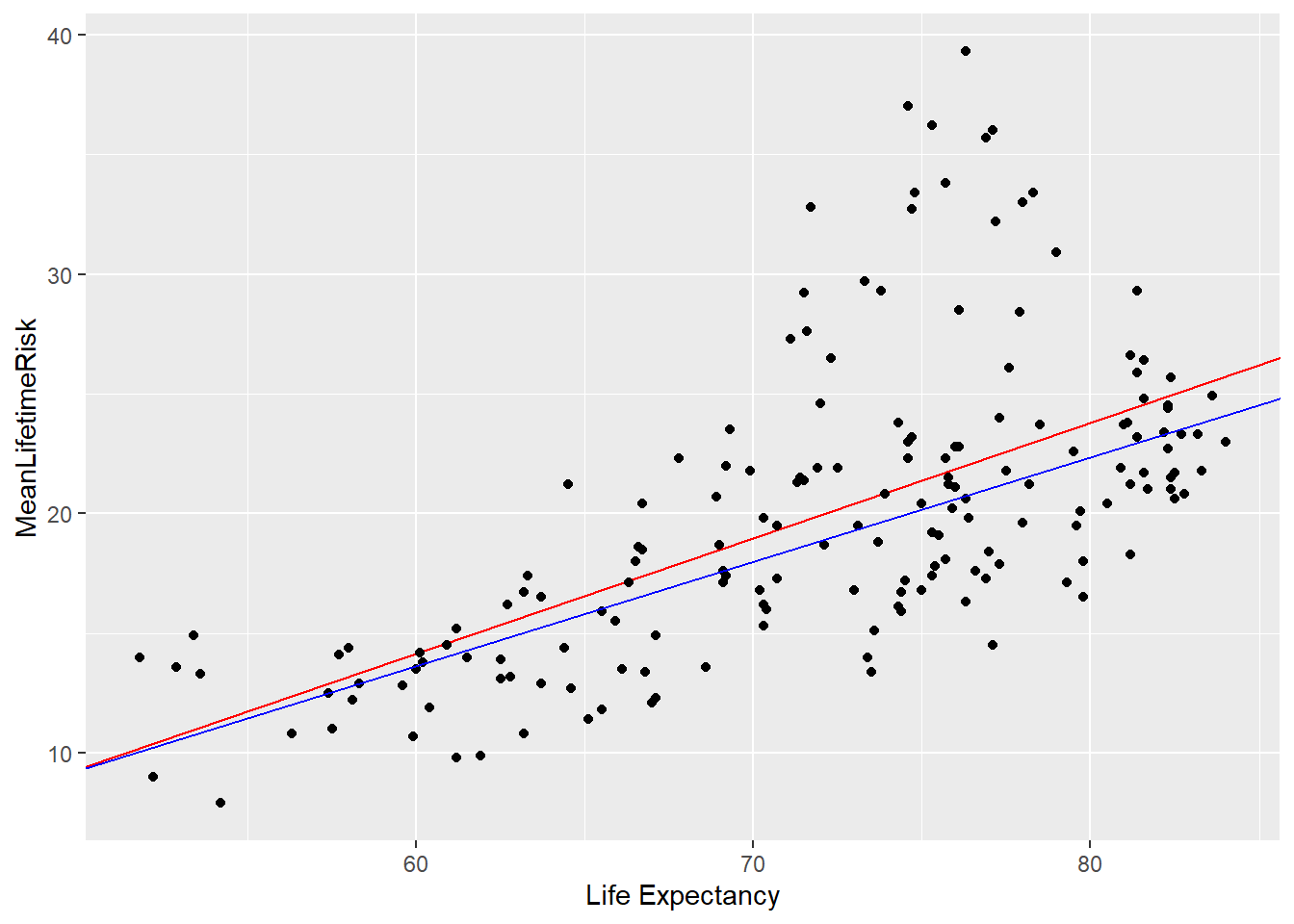
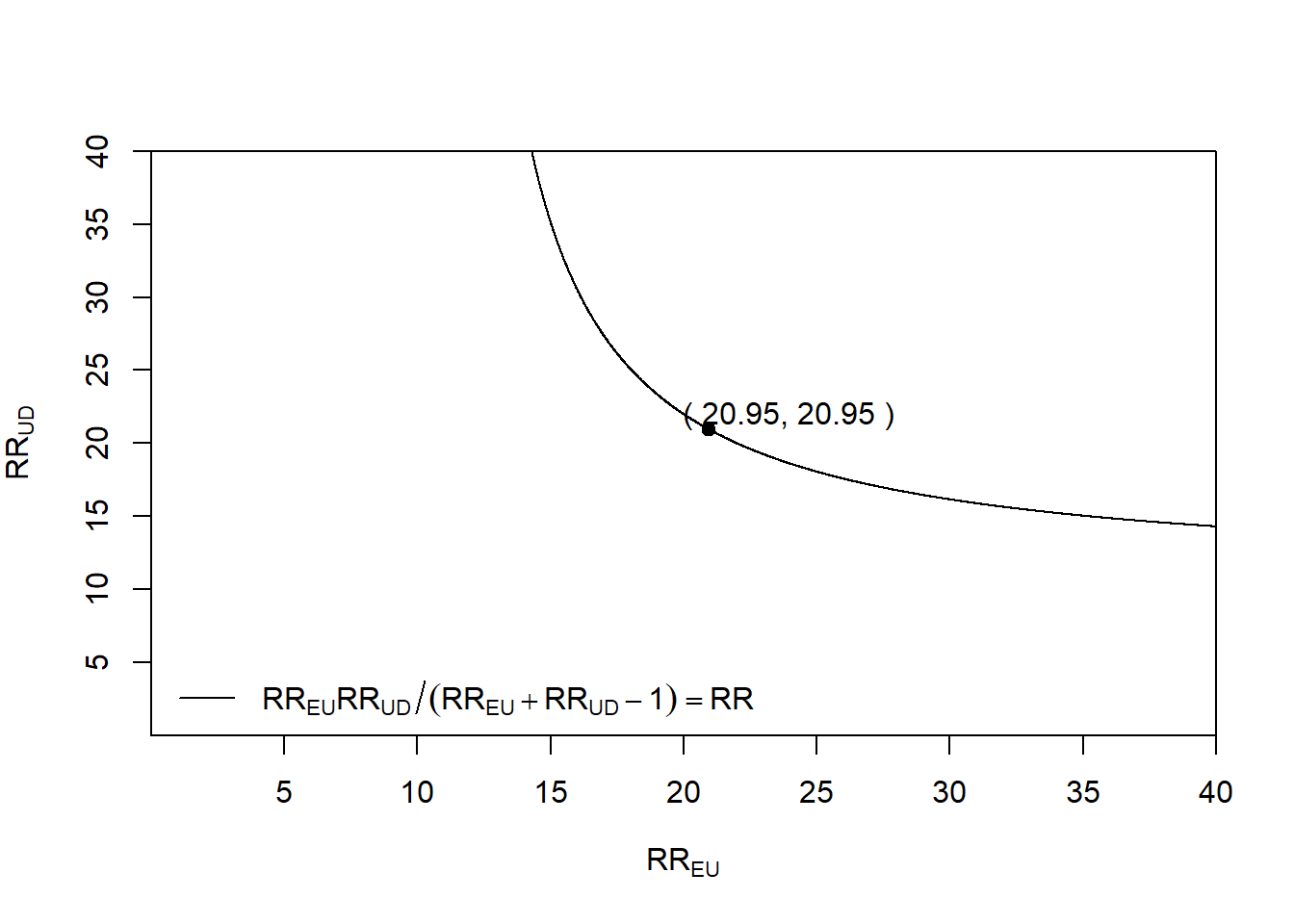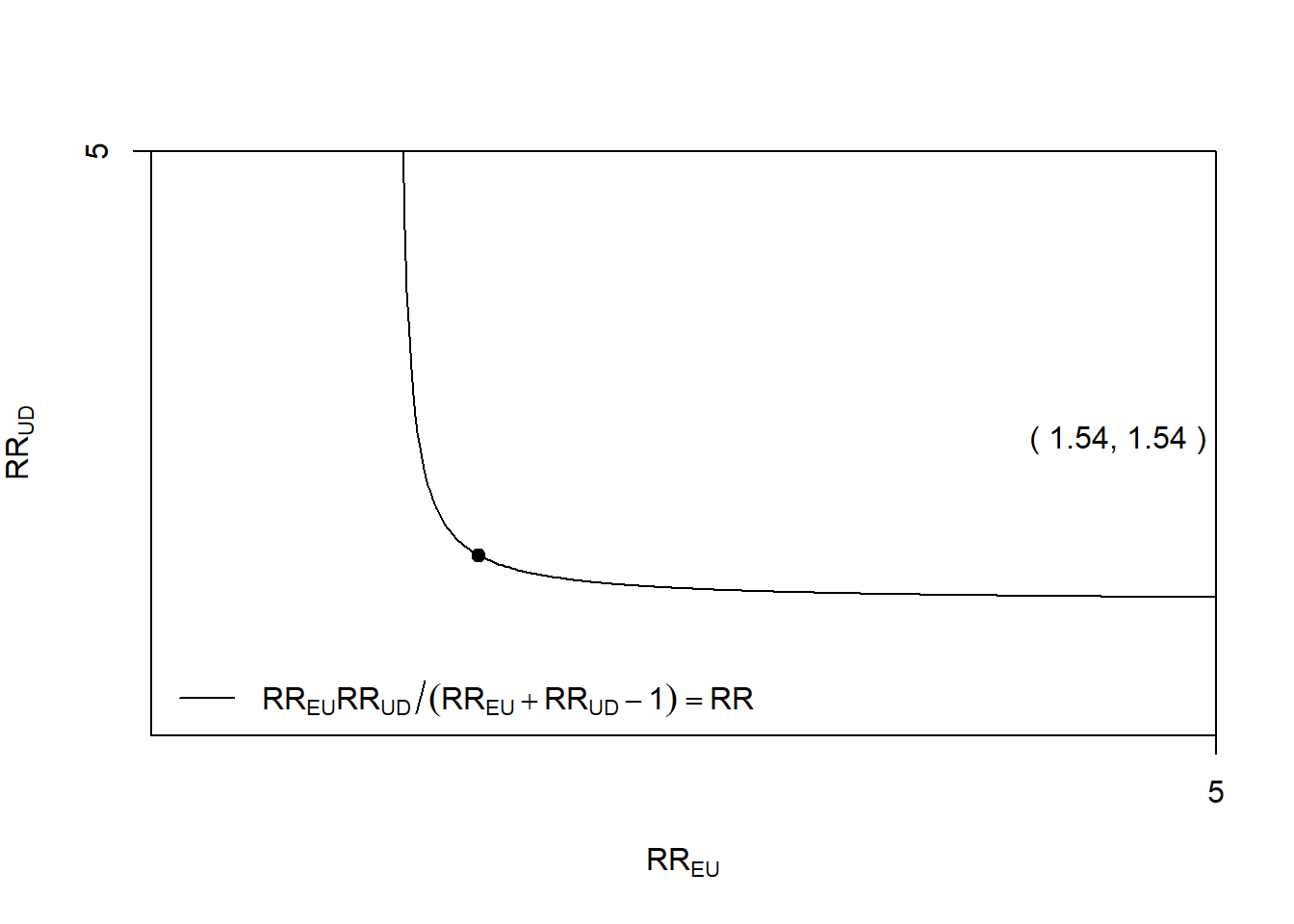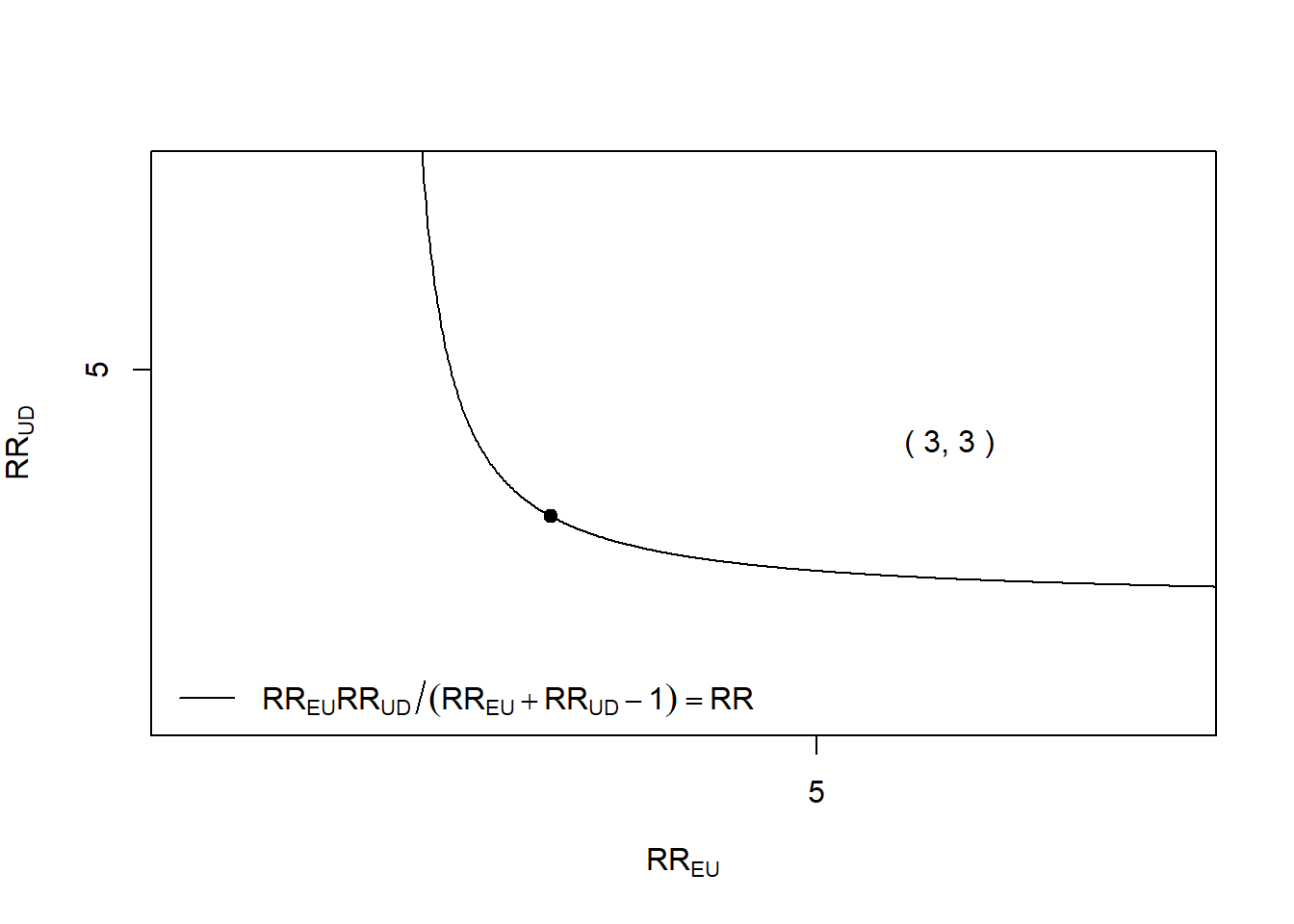Rows: 1,309
Columns: 11
$ survived <fct> no, yes, yes, yes, no, no, no, no, yes, yes, yes, yes, no, no…
$ pclass <ord> 3, 1, 3, 1, 3, 3, 1, 3, 3, 2, 3, 1, 3, 3, 3, 2, 3, 2, 3, 3, 2…
$ name <chr> "Braund, Mr. Owen Harris", "Cumings, Mrs. John Bradley (Flore…
$ sex <fct> male, female, female, female, male, male, male, male, female,…
$ age <dbl> 22, 38, 26, 35, 35, NA, 54, 2, 27, 14, 4, 58, 20, 39, 14, 55,…
$ sib_sp <int> 1, 1, 0, 1, 0, 0, 0, 3, 0, 1, 1, 0, 0, 1, 0, 0, 4, 0, 1, 0, 0…
$ parch <int> 0, 0, 0, 0, 0, 0, 0, 1, 2, 0, 1, 0, 0, 5, 0, 0, 1, 0, 0, 0, 0…
$ ticket <chr> "A/5 21171", "PC 17599", "STON/O2. 3101282", "113803", "37345…
$ fare <dbl> 7.2500, 71.2833, 7.9250, 53.1000, 8.0500, 8.4583, 51.8625, 21…
$ cabin <chr> NA, "C85", NA, "C123", NA, NA, "E46", NA, NA, NA, "G6", "C103…
$ embarked <fct> S, C, S, S, S, Q, S, S, S, C, S, S, S, S, S, S, Q, S, S, C, S…